Description:
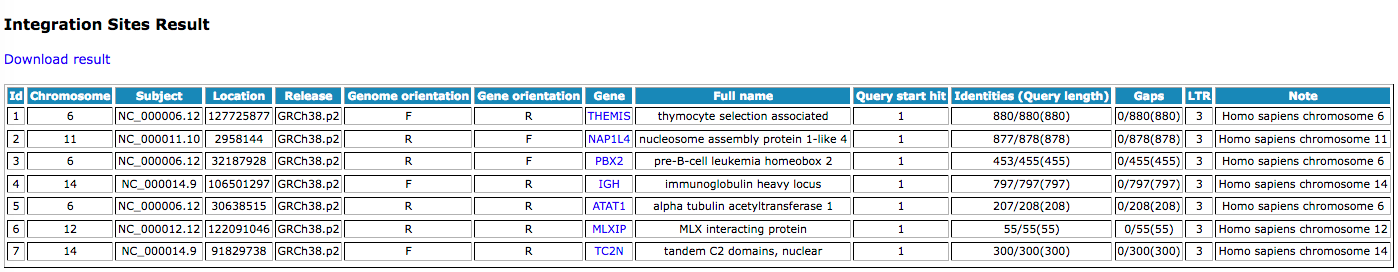
The Integration Sites program developed in the Mullins Lab at the University of Washington is a bioinformatics tool used to detect the integration sites of HIV into the human genome. After the user uploads a FASTA file with the query sequences, the program performs a BLAST search against the human genome (GRCh38.p2 or GRCh37.p5 reference assembly) and outputs the information shown in the example output below. If a sequence is not found in the human genome, the program will then BLAST against the HIV HXB2 sequence. If "Trim LTR sequence first" is checked, the program will look for the sequence of "TCTCTAGCA", a conserved sequence at the 3' end of the LTR, and trim it along with the adjacent viral sequence if the 3'LTR is selected, or the sequence of "GCCCTTCCA" and the adjacent viral sequence will be trimmed if the 5'LTR is selected. For the details how to prepare sequences and run the tool, please read the instruction PDF here.
Sample output:
Contact:
For any questions, bugs and suggestions, please send email to mullspt@uw.edu and include a few sentences describing, briefly, the nature of your questions and include contact information.